Tutorial for HSDatabase
[Download Tutorial for HSDatabase.pdf]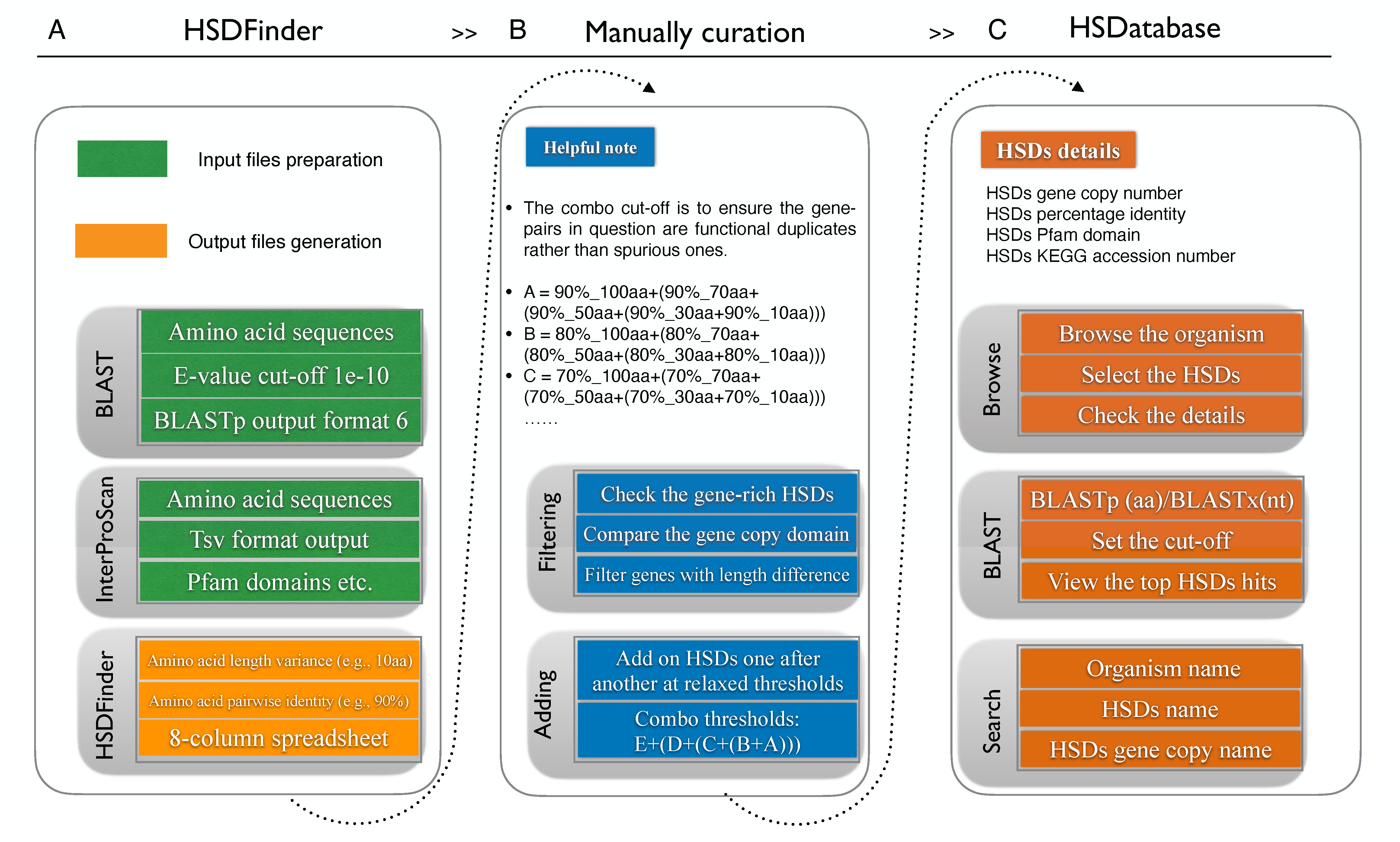
Webinars: Using Bioinformatics Tools to Predict, Collect and Visualize the Highly Similar Duplicates in Eukaryotic Genomes
HSDatabase - a database of highly similar duplicate genes in eukaryotic genomes. (http://hsdfinder.com/database/)
Series 4: How to use the HSDatabase - browse and search?
Series 5: How to use the HSDatabase - BLAST and KEGG?
FAQ
There are many features in HSDatabase including, but not limited to the information of HSDs number, gene copy number and gene copy length. The protein function domains and pathway of HSDs are also collected from database of KEGG and InterProScan. What’s more, a BLAST search option is provided for users to conveniently explore potential homologous sequences of interest. The HSDatabase aims to become a useful platform for the prediction and comparative analysis of HSDs in the eukaryotic genomes of different survival environments, which might deepen our insights into the gene duplication mechanisms driving the genome adaptation.
The predicted HSDs will be manually curated before submitting to the HSDatabase. We filtered possible false positive HSDs with unfit gene copy length and add novel HSDs if necessary, especially, for those satisfy the criteria of near-identical protein lengths (within 10 amino acids and >80% pairwise identities). HSDatabase is built using a relational database (MySQL) allowing rapid retrieval of data and making resource easily maintainable. One entry corresponds to one eukaryote genome. The genomes can be browsed via the categories of plant and animal or the phylogeny for specific genome target. The genome entry is then split into many subcategories of HSDs entries. The database access is via a web interface based on python script and provides various ways to search for HSDs entries, such as organism’s name, HSDs accession number, and gene copy name, etc. HSDatabase can allow users to conveniently BLAST a query to find a homologous match. HSDatabase will be updated timely and the latest version is HSDatabase v1.5.
HSDatabase is a database of categorizing these highly similar duplicate genes (HSDs) in nuclear eukaryotes, which documented total of 117,864 HSDs from 40 eukaryotic species.
The HSDatabase is based on the data provided by the NCBI FTP site. If your species is stored in the FTP site, it will be a helpful to provide us the FTP links to the peptide database. At least, a link to the species information is required. There will be several necessary files before documenting into the database, such as:
- Peptide sequence will be screened by the script isoform2one to remove redundant alternative splicing transcripts (https://github.com/zx0223winner/isoform2one)
- All-against-all BLAST result file
- InterProscan file
- KEGG file
Although there is no golden rule to distinguish partial duplicates from more complete ones, it is believed that the candidate HSDs turn to have at least less than 50% amino acid length difference and similar function of conserved domains.
To balance the HSDs detection sensitivity and accuracy, we have improved the duplicates genes detection and decreased the “snowball effect” via using a series of combo threshold from 90%_10aa to 90%_100aa and from 50%_10aa to 50%_100aa, which can to some extent balance the HSDs detection sensitivity and accuracy. The combo threshold was selected via using a series of thresholds: E + (D + (C + (B +A))).
To balance the HSDs detection sensitivity and accuracy, we have improved the duplicates genes detection and decreased the “snowball effect” via using a series of combo threshold from 90%_10aa to 90%_100aa and from 50%_10aa to 50%_100aa, which can to some extent balance the HSDs detection sensitivity and accuracy. The combo threshold was selected via using a series of thresholds: E + (D + (C + (B +A))).
- A = 90%_100aa+(90%_70aa+(90%_50aa+(90%_30aa+90%_10aa)))
- B = 80%_100aa+(80%_70aa+(80%_50aa+(80%_30aa+80%_10aa)))
- C = 70%_100aa+(70%_70aa+(70%_50aa+(70%_30aa+70%_10aa)))
- D = 60%_100aa+(60%_70aa+(60%_50aa+(60%_30aa+60%_10aa)))
- E = 50%_100aa+(50%_70aa+(50%_50aa+(50%_30aa+50%_10aa)))
The speed of the link to the NCBI genome browser is impacted by the number of genes being viewed, peak Internet hours, and website maintenance. This is true as well for the alignments and %identity data, which uses third-party plugins. We suggest that users open one HSD item at a time. We hope to update the website for speed, etc., as the various third-party tools are improved.
Due to the limitation of the strategy used to collect HSDs, there are some large groups of HSD candidates in the database that likely diverged in function from one another and, thus, are not inducing a gene dosage benefit. In the database, we have labelled these putatively diverged HSD groups as “candidate HSDs” and have added a warning note that users should proceed with caution when working with these datasets.