What's HSDatabase
HSDs has served as a critical database in the eukaryotic genomics, and the database has facilitated the studies associated with the highly similar duplcaites, such as gene duplicates detections, gene duplicates prediction. The predictions of HSDs stored in HSDatabse have been supported by a series of experiments, including those published in New Phytologist, iScience etc..
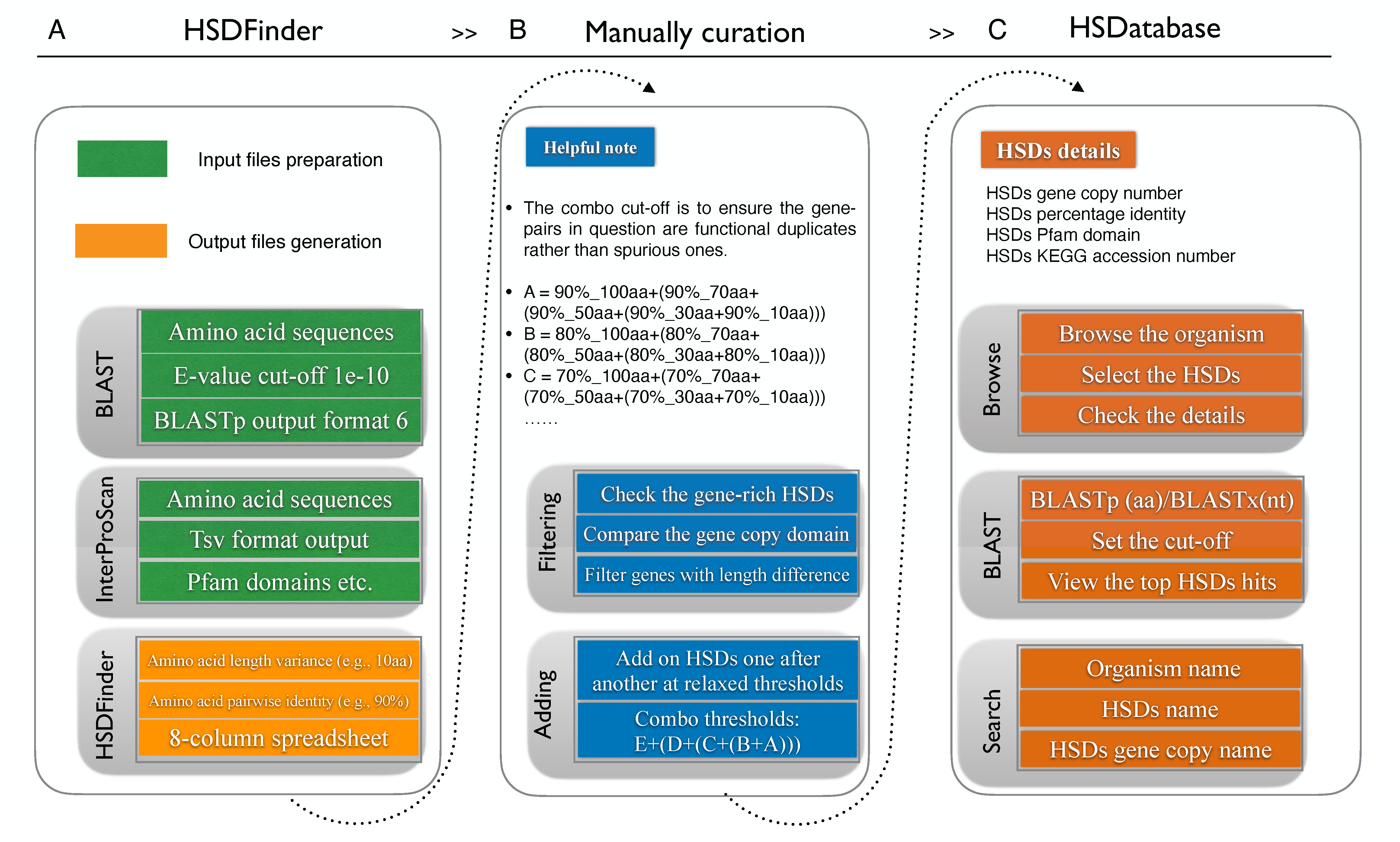
The predicted HSDs in selected eukaryotic nuclear genomes.
Citation and Useful Links
Citation
Xi Zhang, Yining Hu, Zhengyu Cheng, John M. Archibald (2023). HSDecipher: A pipeline for comparative genomic analysis of highly similar duplicate genes in eukaryotic genomes. Star Protocols. doi: https://doi.org/10.1016/j.xpro.2022.102014
Xi Zhang, Yining Hu, David Roy Smith (2022). An overview of online resources for intra-species detection of gene duplications. Frontiers in Genetics. doi: http://doi.org/10.3389/fgene.2022.1012788.
Xi Zhang, Yining Hu, David Roy Smith (2022). HSDatabase - a database of highly similar duplicate genes from plants, animals, and algae. Database. doi: http://doi.org/10.1093/database/baac086.
Xi Zhang, Yining Hu, David Roy Smith (2021). HSDFinder: a BLAST-based strategy to search for highly similar duplicated genes in eukaryotic genomes. Frontiers in Bioinformatics. doi: http://doi.org/10.3389/fbinf.2021.803176
Xi Zhang, Yining Hu, David Roy Smith. (2021). Protocol for HSDFinder: Identifying, annotating, categorizing, and visualizing duplicated genes in eukaryotic genomes. STAR Protocols. DOI:https://doi.org/10.1016/j.xpro.2021.100619
Xi Zhang, Marina Cvetkovska, Rachael Morgan-Kiss, Norman P. A. Hüner, David Roy Smith, (2021). Draft genome sequence of the Antarctic green alga Chlamydomonas sp. UWO241. iScience. https://doi.org/10.1016/j.isci.2021.102084
Links to the InterProScan and KEGG
Pfam 34.0 (March 2021, 19179 entries): https://pfam.xfam.org
InterPro 86.0 (June 2021, 38,913 entries):http://www.ebi.ac.uk/interpro/
KEGG Orthology Database: https://www.genome.jp/kegg/ko.html
InterProscan: https://github.com/ebi-pf-team/interproscan
KEGG : https://www.kegg.jp/kegg/